suppressMessages(library(tidyverse))suppressMessages(library(glue))PRE ="/c/Users/Saideep/Box/imlab-data/data-Github/Daily-Blog-Sai"## COPY THE DATE AND SLUG fields FROM THE HEADERSLUG="implementing-personalized-mutagenesis"## copy the slug from the headerbDATE='2023-03-10'## copy the date from the blog's header hereDATA =glue("{PRE}/{bDATE}-{SLUG}")if(!file.exists(DATA)) system(glue::glue("mkdir {DATA}"))
[1] 1
Code
WORK=DATA
Context
Yesterday I restarted the pipeline after Temi debugged it. After it ran for some time, I checked the results. The results are looking much more reasonable now (values are close to the expected range of values). However, the values themselves are not exactly identical to the ones I generated before, although they seem to correlate across individuals. Temi and I will again take a look at the input sequences to finalize our consensus.
Yesterday, I also formulated personalized mutagenesis experiments. Today I would like to implement the pipeline for running these experiments, and run a subset to help answer: “How closely does do marginal effect sizes match when conditioned on reference background vs. personalized backgrounds?”
Personalized mutagenesis implementation
The first step is to modify the existing merged pipeline so that it also outputs reverse compliment sequences. This involves writing our own function to reverse compliment one-hot-encoded sequence. Temi and I worked on this together because it is good practice at this point to make any changes to the core merged pipeline together and test extensively.
Testing reverse complement of one-hot encoding
Fortunately, the one-hot-encoding is very easy to reverse compliment due to in-built symmetry, requiring just two flips, one on each axis. The code below shows this:
The other thing to consider is that during the quantification we must use “reversed” TSS bins for quantification on the reverse complement input. This is also easily accomplished following:
\(TSSBin_n = TotalBins - 1 - TSSBin_o\)
In other words, we just subtract the original bin index from the total number of bins minus one to get its new index, demo’ed here:
This is now implemented in the quantification steps.
Verification of Kircher Mutagenesis (again)
Previously I had implemented a script to replicate Kircher Mutagenesis. I can now rerun this with the correct merged pipeline and with the reverse compliment sequences.
After running the pipeline, we need to be extra sure that the results are correct. A first pass of quantification showed that the reverse compliment quantifications were exceedingly low. I then doublechecked this in the following notebook, which showed that there is likely an issue with the reverse complement since the results are not mirrored. This intuition was confirmed again by replication in google colab. Those results are positive, shown here:
colab
Temi is a bit busy, so we will fix this issue at a later time. Fortunately, the full GEUVADIS set appears to be running correctly now.
Source Code
---title: "Implementing Personalized Mutagenesis"author: "Saideep Gona"date: "2023-03-10"format: html: code-fold: true code-summary: "Show the code"execute: freeze: true warning: false---```{r}#| label: Set up box storage directorysuppressMessages(library(tidyverse))suppressMessages(library(glue))PRE ="/c/Users/Saideep/Box/imlab-data/data-Github/Daily-Blog-Sai"## COPY THE DATE AND SLUG fields FROM THE HEADERSLUG="implementing-personalized-mutagenesis"## copy the slug from the headerbDATE='2023-03-10'## copy the date from the blog's header hereDATA =glue("{PRE}/{bDATE}-{SLUG}")if(!file.exists(DATA)) system(glue::glue("mkdir {DATA}"))WORK=DATA```# Context- Yesterday I restarted the pipeline after Temi debugged it. After it ran for some time, I checked the results. The results are looking much more reasonable now (values are close to the expected range of values). However, the values themselves are not exactly identical to the ones I generated before, although they seem to correlate across individuals. Temi and I will again take a look at the input sequences to finalize our consensus.- Yesterday, I also formulated personalized mutagenesis experiments. Today I would like to implement the pipeline for running these experiments, and run a subset to help answer: "How closely does do marginal effect sizes match when conditioned on reference background vs. personalized backgrounds?"## Personalized mutagenesis implementationThe first step is to modify the existing merged pipeline so that it also outputs reverse compliment sequences. This involves writing our own function to reverse compliment one-hot-encoded sequence. Temi and I worked on this together because it is good practice at this point to make any changes to the core merged pipeline together and test extensively.### Testing reverse complement of one-hot encodingFortunately, the one-hot-encoding is very easy to reverse compliment due to in-built symmetry, requiring just two flips, one on each axis. The code below shows this:The other thing to consider is that during the quantification we must use "reversed" TSS bins for quantification on the reverse complement input. This is also easily accomplished following:$TSSBin_n = TotalBins - 1 - TSSBin_o$In other words, we just subtract the original bin index from the total number of bins minus one to get its new index, demo'ed here:```{python}number_of_bins =8bins = [0]*number_of_binsoriginal_bin_position =2bins[original_bin_position] ="original bin"print(bins)bins = [0]*number_of_binsnew_bin_position = number_of_bins - original_bin_position -1bins[new_bin_position] ="new bin"print(bins)```This is now implemented in the quantification steps.### Verification of Kircher Mutagenesis (again)Previously I had implemented a script to replicate Kircher Mutagenesis. I can now rerun this with the correct merged pipeline and with the reverse compliment sequences.After running the pipeline, we need to be extra sure that the results are correct. A first pass of quantification showed that the reverse compliment quantifications were exceedingly low. I then doublechecked this in the following [notebook](rc_validation.html), which showed that there is likely an issue with the reverse complement since the results are not mirrored. This intuition was confirmed again by replication in google colab. Those results are positive, shown here: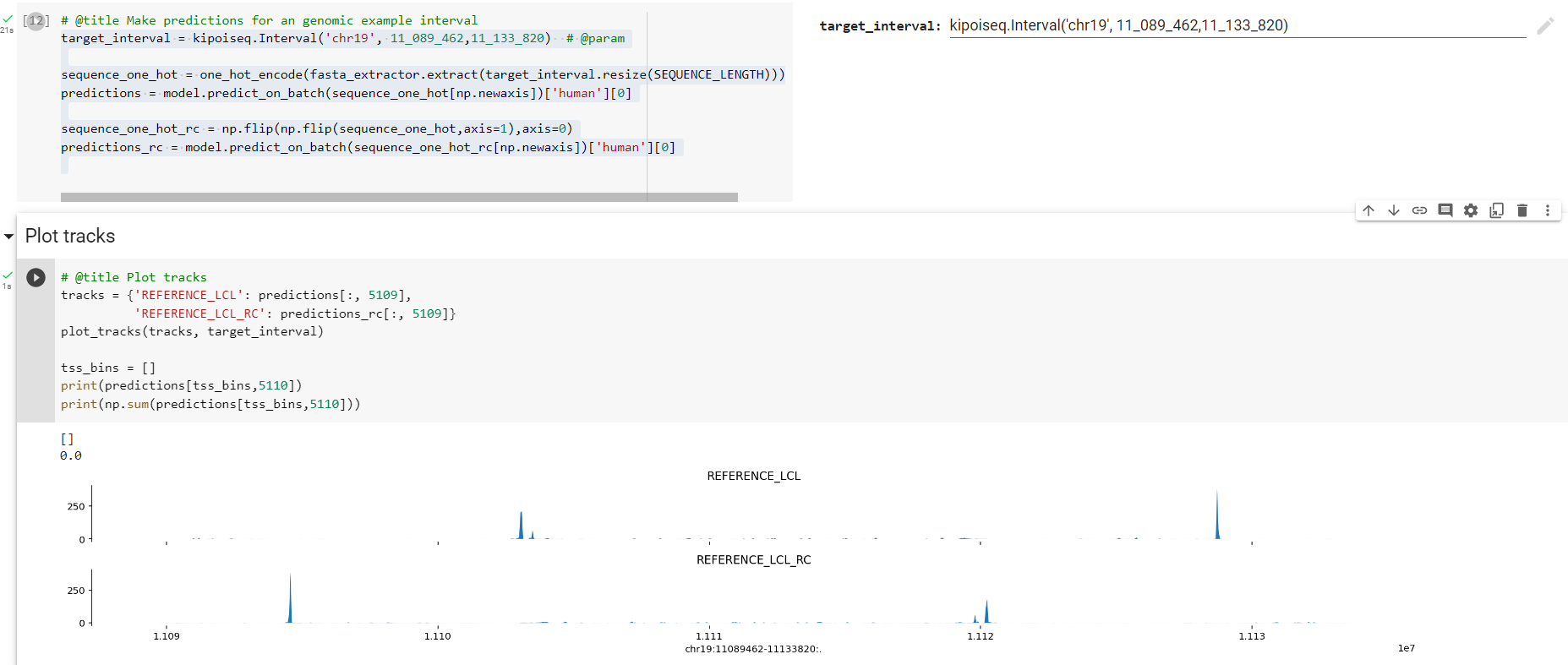Temi is a bit busy, so we will fix this issue at a later time. Fortunately, the full GEUVADIS set appears to be running correctly now.