Code
import h5py
import numpy as np
import torch
import os,sys
import pickle as pkl
import kipoiseq
sys.path.append("./../2023-09-05-ref_to_aracena_mlp")
import ref_to_aracena_modelsSaideep Gona
September 21, 2023
model_type = "cnn"
cur_individual = "AF20"
device = (
"cuda"
if torch.cuda.is_available()
else "mps"
if torch.backends.mps.is_available()
else "cpu"
)
print(device)
if model_type == "linear":
cur_model = ref_to_aracena_models.RefToAracenaMLP(hidden_layer_dims=[])
else:
cur_model = ref_to_aracena_models.RefToAracenaCNN()
cur_model = cur_model.to(device)cpusaved_models_path = "/beagle3/haky/users/saideep/projects/aracena_modeling/saved_models"
trained_model_path = os.path.join(saved_models_path, "/beagle3/haky/users/saideep/projects/aracena_modeling/saved_models/train1_ref_to_aracena_cnn_poissonlog_ep9_lr1e-06.pt")
# trained_model_path = os.path.join(saved_models_path, "train1_ref_to_aracena_cnn_poissonlog_ep19_lr1e-06.pt")
saved_model = torch.load(trained_model_path, map_location=device)
cur_model.load_state_dict(saved_model["model_state_dict"])
cur_model.eval()RefToAracenaCNN(
(model): Sequential(
(0): Sequential(
(0): BatchNorm1d(5313, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(1): GELU()
(2): Conv1d(5313, 24, kernel_size=(4,), stride=(2,), padding=(1,))
(3): BatchNorm1d(24, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(4): GELU()
(5): Conv1d(24, 24, kernel_size=(4,), stride=(2,), padding=(1,))
(6): BatchNorm1d(24, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(7): GELU()
(8): Conv1d(24, 24, kernel_size=(4,), stride=(2,), padding=(1,))
)
(1): Sequential(
(0): Flatten(start_dim=1, end_dim=-1)
(1): Linear(in_features=2688, out_features=10752, bias=True)
(2): Softplus(beta=1, threshold=20)
(3): Unflatten(dim=1, unflattened_size=(12, 896))
)
)
)<Axes: >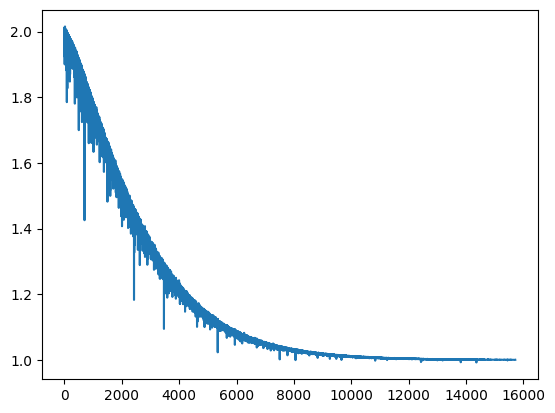
These sites and corresponding TSS sites were extracted in 2023-09-20-diagnose_training_tracks_issue and serve as a good test set to observe model performance.
def plot_tracks(tracks, interval, vlines,height=1.5):
fig, axes = plt.subplots(len(tracks), 1, figsize=(20, height * len(tracks)), sharex=True)
for ax, (title, y) in zip(axes, tracks.items()):
ax.fill_between(np.linspace(interval[1], interval[2], num=len(y)), y)
ax.set_title(title, fontsize=18)
ax.vlines(list(vlines),0, max(y), color="red")
sns.despine(top=True, right=True, bottom=True)
ax.set_xlabel("_".join([str(x) for x in interval]))
plt.tight_layout()
def query_epigenome_from_hdf5(region, individual="AF20"):
import pandas as pd
import h5py
col_region = "_".join([region[0].strip("chr"),region[1],region[2]])
mapping_table = pd.read_csv("/beagle3/haky/users/saideep/projects/aracena_modeling/hdf5_training/index_files/remapped_table_filt.csv", index_col=0)
if col_region not in mapping_table.columns:
print(col_region, "not present in hdf5")
return None,"region not present in hdf5"
query_index = int(mapping_table.loc["ref-epigenome", col_region])
query_group = mapping_table.loc["group", col_region]
print(query_index,query_group)
enformer_ref_targets = f"/beagle3/haky/users/saideep/projects/aracena_modeling/hdf5_training/ref-epigenome_{query_group}_aracena.h5"
with h5py.File(enformer_ref_targets, 'r') as ert:
query = ert['ref_epigenome'][:,:,query_index]
return query, None
def query_aracena_from_hdf5(region, individual="AF20"):
import pandas as pd
import h5py
col_region = "_".join([region[0].strip("chr"),region[1],region[2]])
mapping_table = pd.read_csv("/beagle3/haky/users/saideep/projects/aracena_modeling/hdf5_training/index_files/remapped_table_filt.csv", index_col=0)
if col_region not in mapping_table.columns:
print(col_region, "not present in hdf5")
return None,"region not present in hdf5"
query_index = int(mapping_table.loc[individual, col_region])
query_group = mapping_table.loc["group", col_region]
print(query_index,query_group)
enformer_ara_targets = f"/beagle3/haky/users/saideep/projects/aracena_modeling/hdf5_training/{individual}_{query_group}_aracena.h5"
print(enformer_ara_targets)
with h5py.File(enformer_ara_targets, 'r') as ert:
query = ert['targets'][:,0,query_index]
return query, None
def predict_on_ref_epi(cur_model, ref_epi_input, model_type, device = "cpu"):
cur_model = cur_model.to(device)
if model_type == "linear":
input_tensor = torch.from_numpy(ref_epi_input).to(device).unsqueeze(0)
elif model_type == "cnn":
print("pred_func", ref_epi_input.shape)
input_tensor = torch.from_numpy(ref_epi_input).to(device).unsqueeze(0).swapaxes(1,2)
prediction_raw = cur_model(input_tensor)
if model_type == "linear":
prediction = prediction_raw.squeeze().cpu().detach().numpy()[:,:].swapaxes(0,1)
elif model_type == "cnn":
print("pred_func", prediction_raw.shape)
prediction = prediction_raw.squeeze().cpu().detach().numpy()[:,:].swapaxes(0,1)
print("pred_func_last",prediction.shape)
return predictionwith open("/beagle3/haky/users/saideep/github_repos/Daily-Blog-Sai/posts/2023-09-20-diagnose_training_tracks_issue/tss_mapping_dict.pkl", "rb") as tmd:
tss_mapping_dict = pkl.load(tmd)
with open("/beagle3/haky/users/saideep/github_repos/Daily-Blog-Sai/posts/2023-09-20-diagnose_training_tracks_issue/tss_mapping_dict_gene_names.pkl", "rb") as tmd:
tss_mapping_dict_gene_names = pkl.load(tmd)
c=0
for region in tss_mapping_dict.keys():
print(region)
if c < 3:
c+=1
continue
if c==10:
break
else:
c+=1
region_split = region.split("_")
print("collecting ref query")
ref_query_hdf5, flag = query_epigenome_from_hdf5(region_split)
if flag == "region not present in hdf5":
continue
ara_query_hdf5, flag = query_aracena_from_hdf5(region_split)
if flag == "region not present in hdf5":
continue
region_for_plot_raw = [region_split[0],int(region_split[1]),int(region_split[2])]
region_for_plot_kipoi_resized = kipoiseq.Interval(*region_for_plot_raw).resize(896*128)
region_for_plot = [region_for_plot_kipoi_resized.chrom,
region_for_plot_kipoi_resized.start,
region_for_plot_kipoi_resized.end]
region_browser = region_for_plot[0]+":"+str(region_for_plot[1])+"-"+str(region_for_plot[2])
print("ref_query",ref_query_hdf5.shape)
model_prediction = predict_on_ref_epi(cur_model, ref_query_hdf5, "cnn")
print(region_browser, "genes:", tss_mapping_dict_gene_names[region], "region len:", (int(region_for_plot[2])-int(region_for_plot[1])))
tracks_for_plot = {"ref_epi_hdf5":ref_query_hdf5[:,4766],
"aracena_hdf5":ara_query_hdf5,
"model_predict":model_prediction[:,0]}
plot_tracks(tracks_for_plot,region_for_plot, tss_mapping_dict[region])chr4_113630947_113762019
chr11_18427720_18558792
chr16_85805681_85936753
chr7_136783551_136914623
collecting ref query
7_136783551_136914623 not present in hdf5
chr16_24521594_24652666
collecting ref query
7 train1
8 train1
/beagle3/haky/users/saideep/projects/aracena_modeling/hdf5_training/AF20_train1_aracena.h5
ref_query (896, 5313)
pred_func (896, 5313)
pred_func torch.Size([1, 12, 896])
pred_func_last (896, 12)
chr16:24529786-24644474 genes: ['RBBP6'] region len: 114688
chr17_74167260_74298332
collecting ref query
10 train1
11 train1
/beagle3/haky/users/saideep/projects/aracena_modeling/hdf5_training/AF20_train1_aracena.h5
ref_query (896, 5313)
pred_func (896, 5313)
pred_func torch.Size([1, 12, 896])
pred_func_last (896, 12)
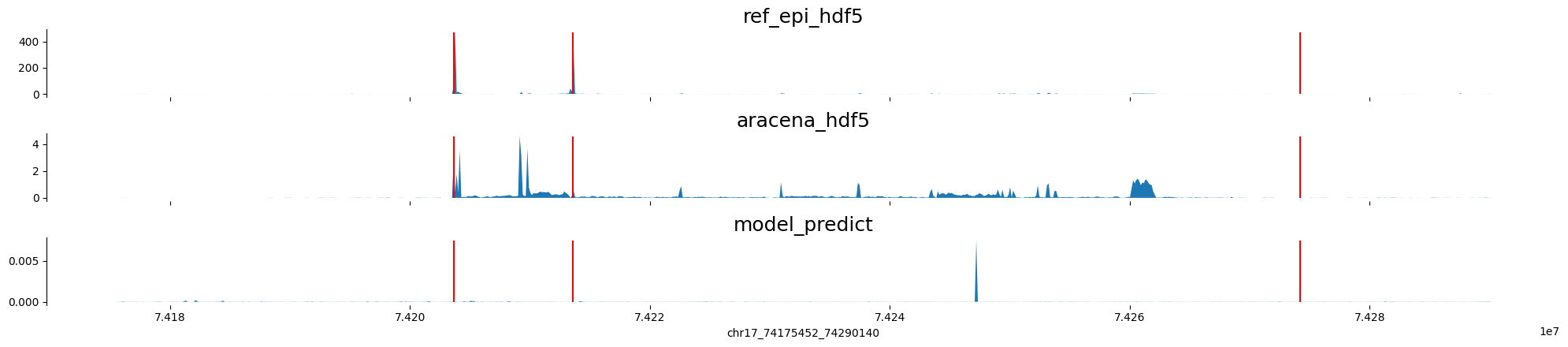
chr17:74175452-74290140 genes: ['RPL38', 'TTYH2', 'DNAI2'] region len: 114688
chr9_93148172_93279244
collecting ref query
13 train1
14 train1
/beagle3/haky/users/saideep/projects/aracena_modeling/hdf5_training/AF20_train1_aracena.h5
ref_query (896, 5313)
pred_func (896, 5313)
pred_func torch.Size([1, 12, 896])
pred_func_last (896, 12)
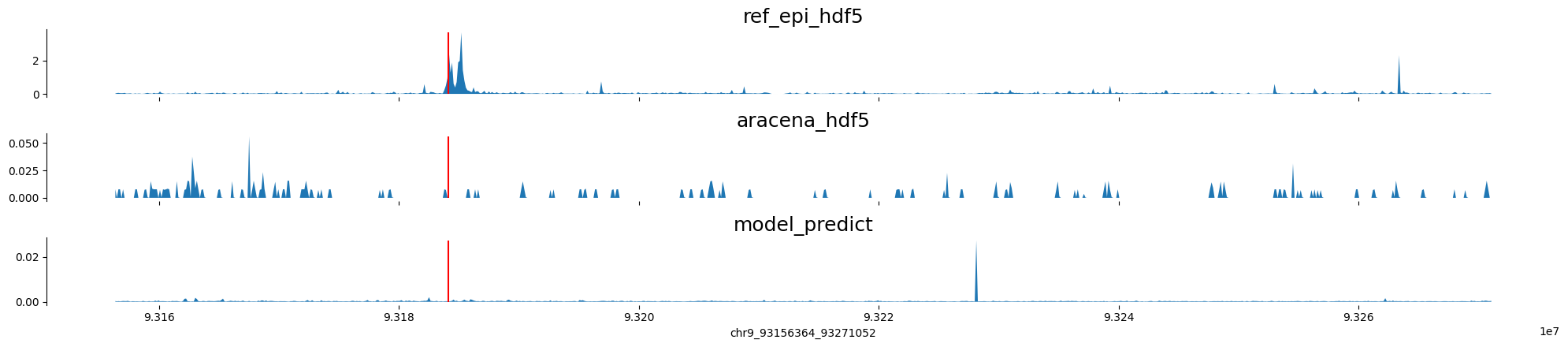
chr9:93156364-93271052 genes: ['WNK2'] region len: 114688
chr4_165060563_165191635
collecting ref query
17 train1
18 train1
/beagle3/haky/users/saideep/projects/aracena_modeling/hdf5_training/AF20_train1_aracena.h5
ref_query (896, 5313)
pred_func (896, 5313)
pred_func torch.Size([1, 12, 896])
pred_func_last (896, 12)
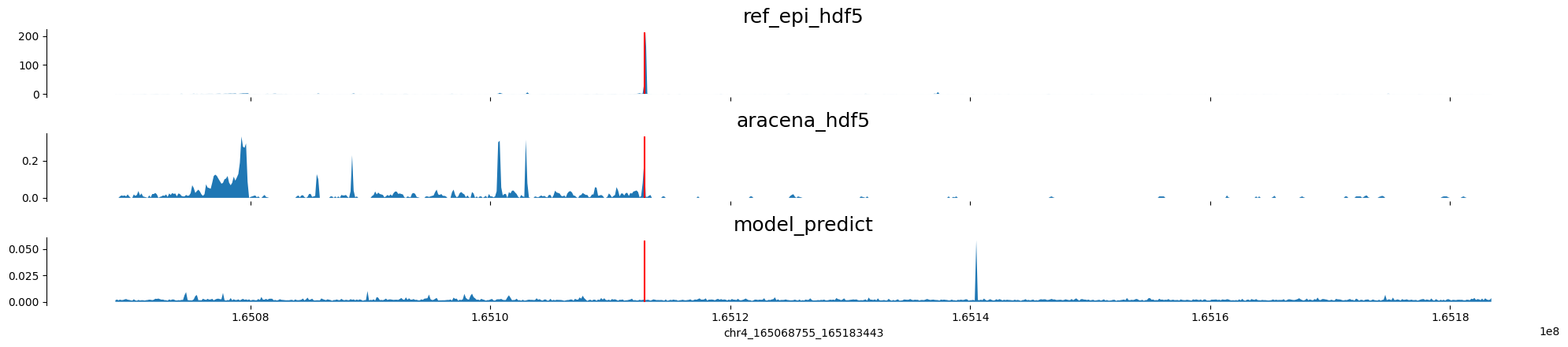
chr4:165068755-165183443 genes: ['TMEM192'] region len: 114688
chr17_19514821_19645893
collecting ref query
18 train1
19 train1
/beagle3/haky/users/saideep/projects/aracena_modeling/hdf5_training/AF20_train1_aracena.h5
ref_query (896, 5313)
pred_func (896, 5313)
pred_func torch.Size([1, 12, 896])
pred_func_last (896, 12)
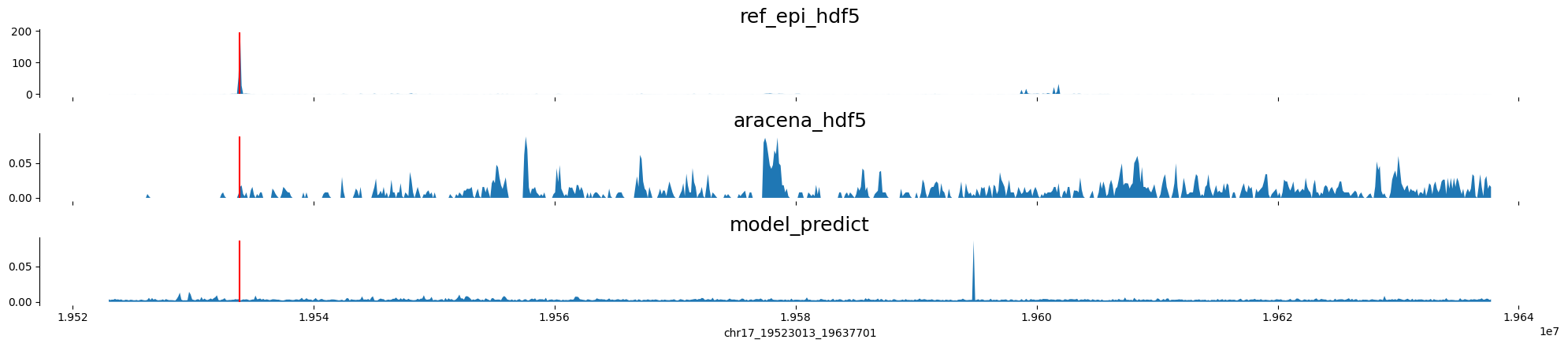
chr17:19523013-19637701 genes: ['SLC47A1'] region len: 114688
chr3_52196061_52327133
collecting ref query
19 train1
20 train1
/beagle3/haky/users/saideep/projects/aracena_modeling/hdf5_training/AF20_train1_aracena.h5
ref_query (896, 5313)
pred_func (896, 5313)
pred_func torch.Size([1, 12, 896])
pred_func_last (896, 12)
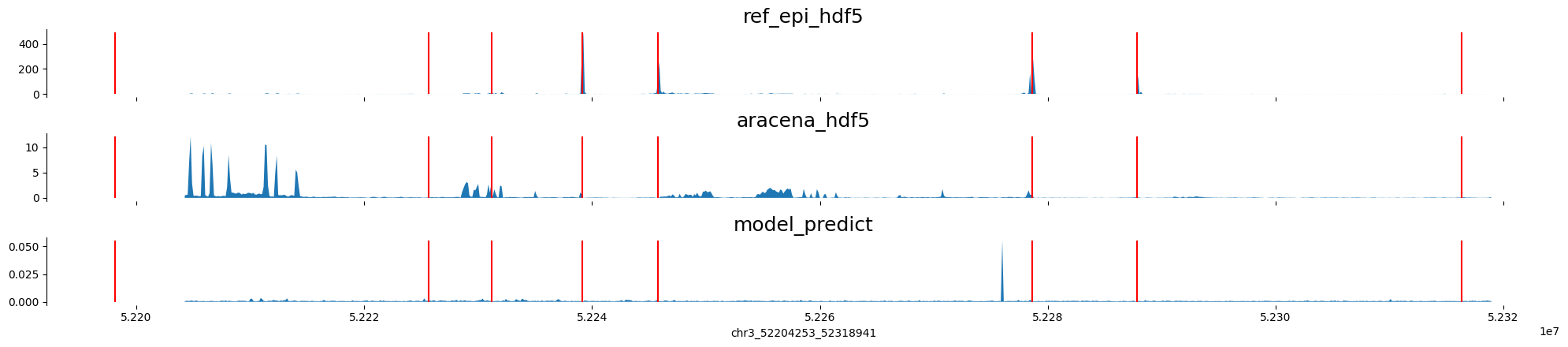
chr3:52204253-52318941 genes: ['ALAS1', '', 'TLR9', 'TWF2', 'PPM1M', 'WDR82', 'GLYCTK', 'DNAH1'] region len: 114688
chr2_167285886_167416958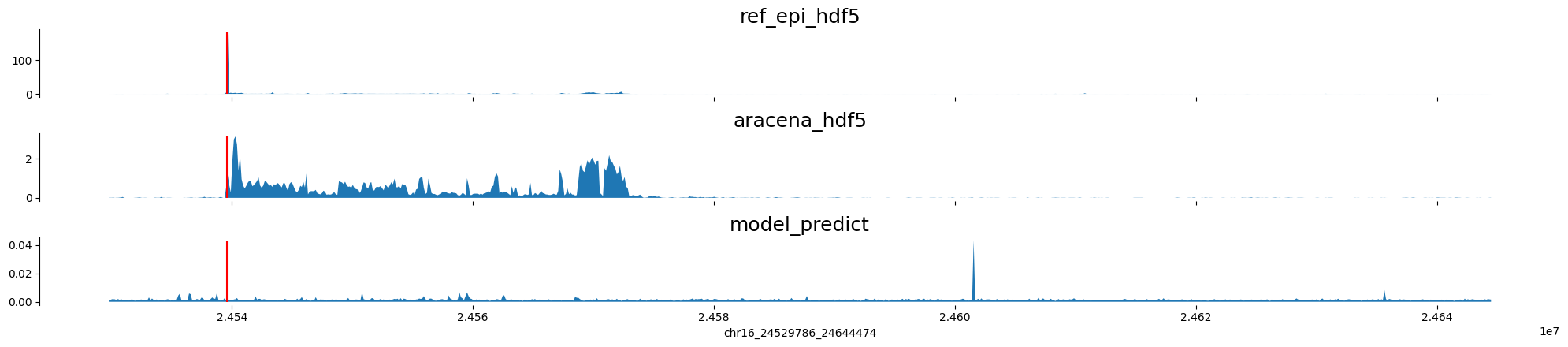
Load in some validation datasets for evaluation
validation_path = "/beagle3/haky/users/saideep/projects/aracena_modeling/hdf5_training/AF20_valid_aracena.h5"
sequence_ind = 40
track_ind = 1
with h5py.File(validation_path, "r") as f:
example_valid = f['targets'][:,:,sequence_ind]
example_valid_reg = f['regions'][:,:,sequence_ind]
print(example_valid.shape)
print(example_valid_reg.shape)
print(example_valid_reg[0,2]-example_valid_reg[0,1])
bigwig_track_readouts = [
"RNAseq",
"ATACseq",
"H3K4me1",
"H3K4me3",
"H3K27ac",
"H3K27me3"
]
conditions = ["Flu","NI"]
bigwig_tracks = []
for condition in conditions:
for bigwig_track_readout in bigwig_track_readouts:
bigwig_tracks.append(condition+"_"+bigwig_track_readout)
(896, 12)
(1, 3)
131072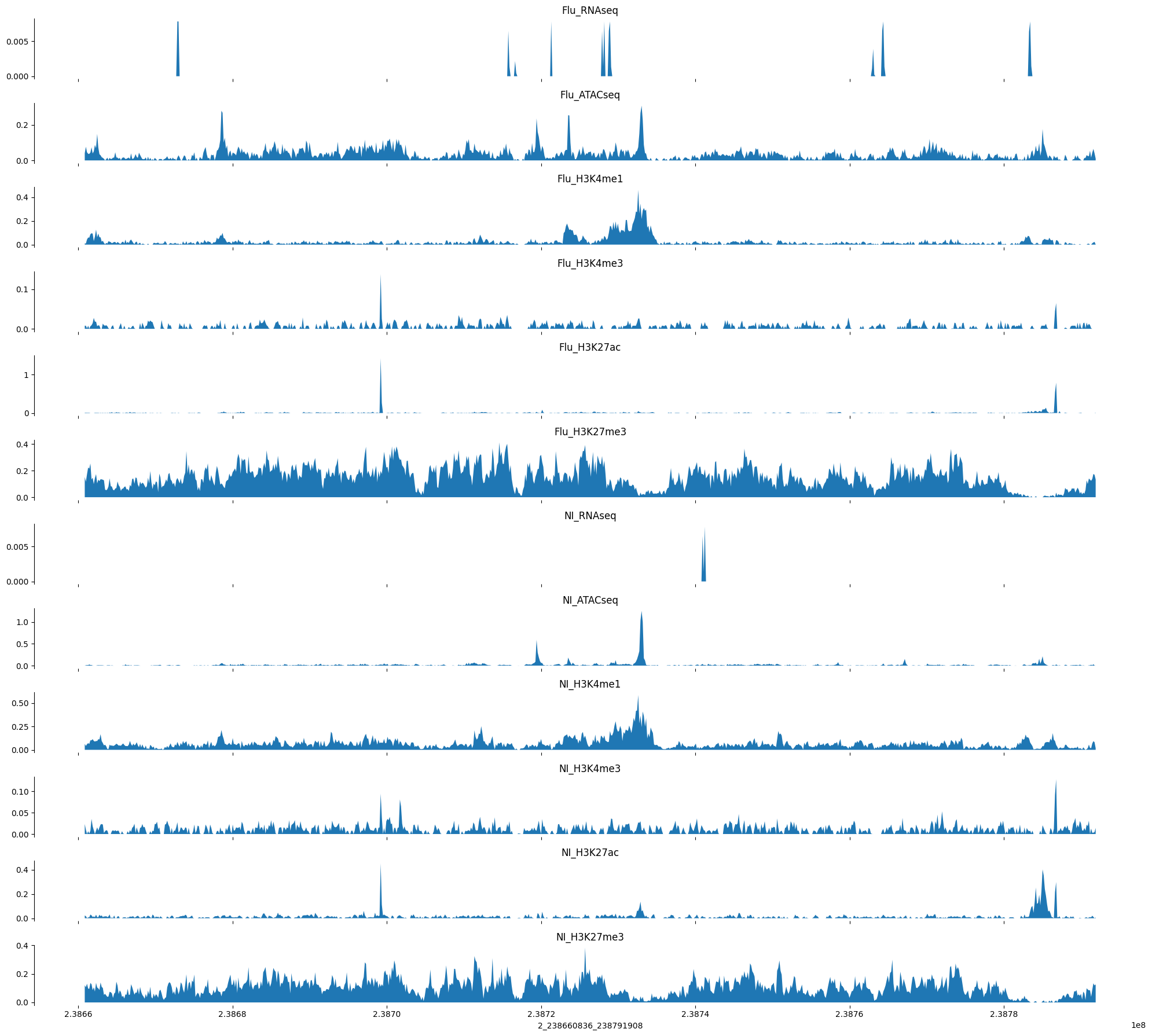
We can now make a prediction on the corresponding input region using our stored model(s)
(9, 1572)target_interval_str = "_".join([str(x) for x in target_interval])
print(target_interval_str)
ref_index = mapping_table.loc["ref-epigenome", target_interval_str]
print(ref_index)
print(target_interval_str in mapping_table.columns)
with h5py.File(f"/beagle3/haky/users/saideep/projects/aracena_modeling/hdf5_training/ref-epigenome_{group}_aracena.h5", "r") as f:
example_ref_pred = f['ref_epigenome'][:,:,int(ref_index)]
example_ref_region = f['regions'][:,:,int(ref_index)]
example_ref_region_str = "_".join([str(x) for x in example_ref_region[0,:]])
print(example_ref_region_str)
print(target_interval_str)
print(example_ref_pred.shape)2_238660836_238791908KeyError: '2_238660836_238791908'def create_tracks(aracena, predictions, track_names):
tracks = {}
for i,track_name in enumerate(track_names):
for track_type in ['aracena', 'model_prediction']:
if track_type == 'aracena':
tracks[track_name+"_"+track_type] = aracena[:,i]
elif track_type == 'model_prediction':
tracks[track_name+"_"+track_type] = predictions[i,:]
return tracks
cur_model = cur_model.to(device)
if model_type == "linear":
input_tensor = torch.from_numpy(example_ref_pred).to(device).unsqueeze(0).to(device)
elif model_type == "cnn":
input_tensor = torch.from_numpy(example_ref_pred).to(device).unsqueeze(0).to(device).swapaxes(1,2)
# input_tensor = torch.from_numpy(example_ref_pred).to(device).unsqueeze(0).to(device)
ex_out_raw = cur_model(input_tensor)
if model_type == "linear":
ex_out = ex_out_raw.squeeze().cpu().detach().numpy()[:,:].swapaxes(0,1)
elif model_type == "cnn":
ex_out = ex_out_raw.squeeze().cpu().detach().numpy()[:,:].swapaxes(0,1)
print(ex_out.shape)
print(np.sum(ex_out, axis=0))
tracks = create_tracks(example_valid, ex_out.swapaxes(0,1), bigwig_tracks)NameError: name 'example_ref_pred' is not definedOK, for this example things are not so promising. It seems like the model was converging on the training set at least, maybe some of those predictions will show promise.
model_type = "cnn"
group = "train1"
cur_individual = "AF20"
device = (
"cuda"
if torch.cuda.is_available()
else "mps"
if torch.backends.mps.is_available()
else "cpu"
)
print(device)
if model_type == "linear":
cur_model = ref_to_aracena_models.RefToAracenaMLP(hidden_layer_dims=[])
else:
cur_model = ref_to_aracena_models.RefToAracenaCNN()
cur_model = cur_model.to(device)
train_path = f"/beagle3/haky/users/saideep/projects/aracena_modeling/hdf5_training/AF20_{group}_aracena.h5"
saved_models_path = "/beagle3/haky/users/saideep/projects/aracena_modeling/saved_models"
trained_model_path = os.path.join(saved_models_path, "/beagle3/haky/users/saideep/projects/aracena_modeling/saved_models/train1_ref_to_aracena_cnn_poissonlog_ep9_lr1e-06.pt")
# trained_model_path = os.path.join(saved_models_path, "train1_ref_to_aracena_cnn_poissonlog_ep19_lr1e-06.pt")
saved_model = torch.load(trained_model_path, map_location=device)
cur_model.load_state_dict(saved_model["model_state_dict"])
cur_model.eval()
sequence_ind = 6
with h5py.File(train_path, "r") as f:
example_train = f['targets'][:,:,sequence_ind]
example_train_reg = f['regions'][:,:,sequence_ind]
target_interval = [str(example_train_reg[0,0]), example_train_reg[0,1], example_train_reg[0,2]]
target_interval_str = "_".join([str(x) for x in target_interval])
print(target_interval_str)
ref_index = mapping_table.loc["ref-epigenome", target_interval_str]
print(ref_index)
with h5py.File(f"/beagle3/haky/users/saideep/projects/aracena_modeling/hdf5_training/ref-epigenome_{group}_aracena.h5", "r") as f:
example_ref_pred = f['ref_epigenome'][:,:,int(ref_index)]
example_ref_region = f['regions'][:,:,int(ref_index)]
print(example_train_reg, example_ref_region)
if model_type == "linear":
input_tensor = torch.from_numpy(example_ref_pred).to(device).unsqueeze(0).to(device)
elif model_type == "cnn":
input_tensor = torch.from_numpy(example_ref_pred).to(device).unsqueeze(0).to(device).swapaxes(1,2)
ex_out_raw = cur_model(input_tensor)
ex_out = ex_out_raw.squeeze().cpu().detach().numpy()[:,:].swapaxes(0,1)
if model_type == "linear":
pass
elif model_type == "cnn":
ex_out = ex_out.swapaxes(0,1)
tracks = create_tracks(example_train, ex_out, bigwig_tracks)
tracks["CAGE_ref"] = example_ref_pred[:,4766]cpu
8_132158314_132289386
5
[[ 8 132158314 132289386]] [[ 8 132158314 132289386]]