Code
import os,sys
import numpy as np
import matplotlib.pyplot as plt
import pandas as pd
import json
import subprocessSaideep Gona
February 28, 2024
I need to test variable input window size for Con-EnPACT modeling.
Runtime: ~30 minutes
path_to_config_template = "/beagle3/haky/users/saideep/github_repos/Con-EnPACT/example_json/flu_4bin_1milscaling.json"
path_to_run_template = "/beagle3/haky/users/saideep/github_repos/Con-EnPACT/run_training.sbatch"
path_to_project_dir = "/beagle3/haky/users/saideep/projects/Con_EnPACT/models"
normalization_strategies = {
"_4_4_lnormTRUE_beforeTRUE_scale1"
# "_3_7_lnormTRUE_beforeTRUE_scale1",
# "_3_7_lnormTRUE_beforeFALSE_scale1",
# "_3_7_lnormFALSE_beforeFALSE_scale1_sourcekallisto"
}
contexts = {"Flu":"/beagle3/haky/users/saideep/projects/aracena_modeling/Inputs/counts_matrices/final_testing/fully_preprocessed_flu_expression_for_predictDBNORMSTRAT.txt",
"NI":"/beagle3/haky/users/saideep/projects/aracena_modeling/Inputs/counts_matrices/final_testing/fully_preprocessed_NI_expression_for_predictDBNORMSTRAT.txt"}
window_sizes = [4]
for normstrat in normalization_strategies:
# if normstrat != "_3_7_lnormFALSE_beforeFALSE_scale1_sourcekallisto":
# continue
for context in contexts.keys():
for window_size in window_sizes:
cur_proj_dir = os.path.join(path_to_project_dir,context+"_ws"+str(window_size)+"_ns"+normstrat)
os.makedirs(cur_proj_dir,exist_ok=True)
with open(path_to_config_template,"r") as f:
config = json.load(f)
config["general_parameters"]["project_directory"] = cur_proj_dir
config["general_parameters"]["context"] = context
config["generate_enpact_training_data"]["input_files"]["normalized_expression_data"] = contexts[context].replace("NORMSTRAT",normstrat)
config["generate_enpact_training_data"]["reference_epigenome"]["num_bins"] = window_size
with open(os.path.join(cur_proj_dir,"config.json"),"w") as f:
json.dump(config,f, indent=4)
with open(path_to_run_template,"r") as f:
run = f.read()
run = run.replace("CONFIG_FILE",os.path.join(cur_proj_dir,"config.json"))
run = run.replace("JOBNAME",context+"_ws"+str(window_size))
run = run.replace("ERROR_LOG", os.path.join(cur_proj_dir,"error_training.log"))
run = run.replace("OUTPUT_LOG", os.path.join(cur_proj_dir,"output_training.log"))
with open(os.path.join(cur_proj_dir,"run_training.sbatch"),"w") as f:
f.write(run)
subprocess.run(["sbatch",
os.path.join(cur_proj_dir,"run_training.sbatch")],
cwd=cur_proj_dir)Submitted batch job 18641516
Submitted batch job 18641517corr_tables = []
for normstrat in normalization_strategies:
for context in contexts.keys():
for window_size in window_sizes:
cur_proj_dir = os.path.join(path_to_project_dir,context+"_ws"+str(window_size)+"_ns"+normstrat)
cur_corr_table = pd.read_csv(os.path.join(cur_proj_dir,"intermediates","evaluate_training","corrs_df_for_subsets.tsv"), sep="\t")
cur_corr_table["normstrat"] = normstrat
cur_corr_table["context"] = context
cur_corr_table["window_size"] = window_size
corr_tables.append(cur_corr_table)
corr_table_all = pd.concat(corr_tables)
corr_table_all.head()| data_subset | cor_id | cor_type | corrs | normstrat | context | window_size | |
|---|---|---|---|---|---|---|---|
| 0 | train | EnPACT_pred_vs_ground_truth | spearman | 0.848034 | _3_7_lnormFALSE_beforeFALSE_scale1_sourcekallisto | Flu | 2 |
| 1 | train | log2_epi_ref_track_vs_ground_truth | spearman | 0.771574 | _3_7_lnormFALSE_beforeFALSE_scale1_sourcekallisto | Flu | 2 |
| 2 | train | EnPACT_pred_vs_ground_truth | pearson | 0.840772 | _3_7_lnormFALSE_beforeFALSE_scale1_sourcekallisto | Flu | 2 |
| 3 | train | log2_epi_ref_track_vs_ground_truth | pearson | 0.586671 | _3_7_lnormFALSE_beforeFALSE_scale1_sourcekallisto | Flu | 2 |
| 4 | valid | EnPACT_pred_vs_ground_truth | spearman | 0.831945 | _3_7_lnormFALSE_beforeFALSE_scale1_sourcekallisto | Flu | 2 |
import seaborn as sns
sns.set_theme(style="whitegrid")
corr_table_enpact = corr_table_all[corr_table_all["cor_id"]=="EnPACT_pred_vs_ground_truth"]
corr_table_enpact["normstrat_short"] = corr_table_enpact["normstrat"].str.replace("_3_7_","").str.replace("_scale1","")
min_corr = corr_table_enpact["corrs"].min() - 0.1
max_corr = corr_table_enpact["corrs"].max() + 0.1
for data_subset in ["train","valid","test"]:
corr_table_enpact_subset = corr_table_enpact[corr_table_enpact["data_subset"]==data_subset]
fg = sns.FacetGrid(data=corr_table_enpact_subset, col="context", row="normstrat_short", margin_titles=True, height=4, aspect=1)
fg.map_dataframe(sns.scatterplot, x="window_size", y="corrs", hue="cor_type", palette="deep").add_legend().set(ylim=(min_corr,max_corr))
fg.figure.suptitle(data_subset, y=1.05)
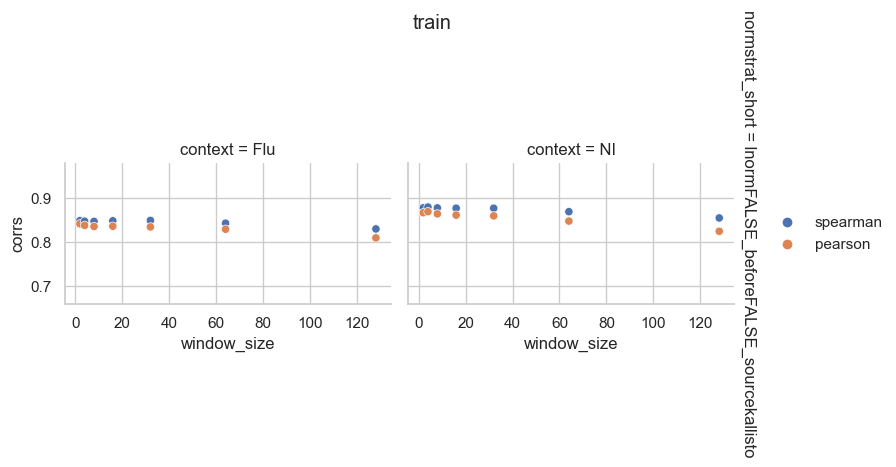
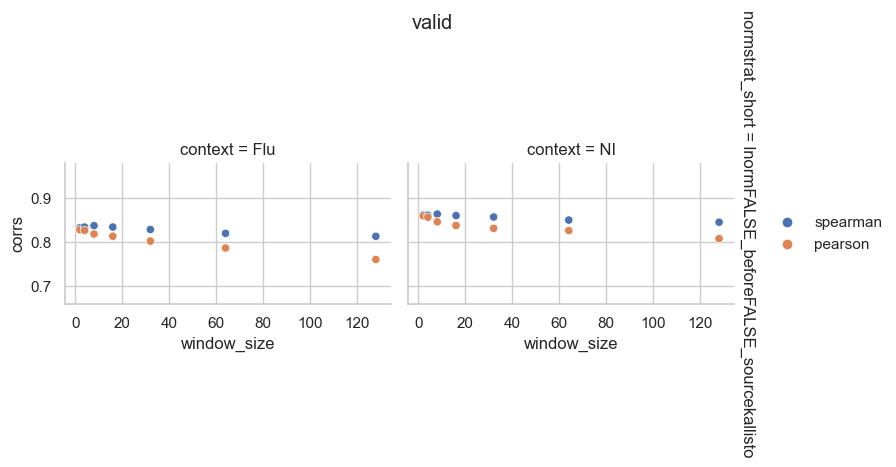
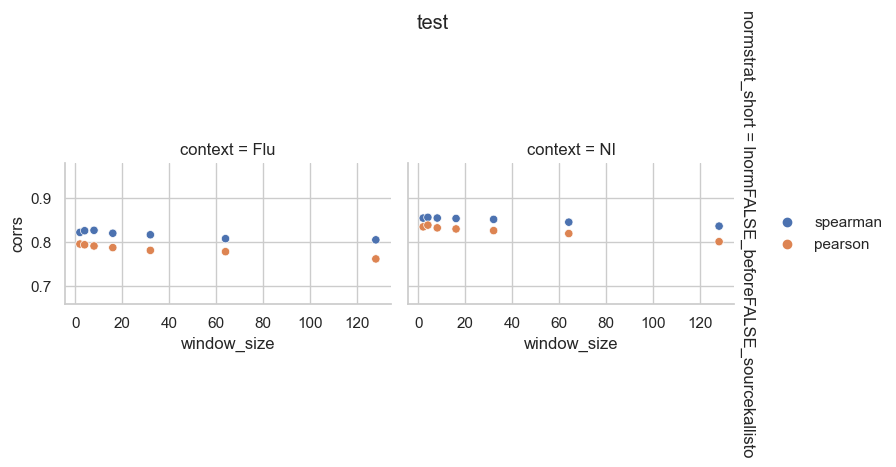
From the perspective of mean expression prediction on the validation dataset, it seems like gene length normalization before TMM normalization is more easily predicted than length normalization after TMM normalization. It also doesn’t seem like window size has a huge effect on the prediction accuracy.
Runtime ~ 8-10 hours per EnPACT model
path_to_config_template = "/beagle3/haky/users/saideep/github_repos/Con-EnPACT/example_json/flu_4bin_1milscaling.json"
path_to_run_linearization_template = "/beagle3/haky/users/saideep/github_repos/Con-EnPACT/run_linearization.sbatch"
path_to_project_dir = "/beagle3/haky/users/saideep/projects/Con_EnPACT/models"
normalization_strategies = {
"_4_4_lnormTRUE_beforeTRUE_scale1"
# "_3_7_lnormTRUE_beforeTRUE_scale1"
# "_3_7_lnormFALSE_beforeFALSE_scale1_sourcekallisto"
}
contexts = {"Flu":"/beagle3/haky/users/saideep/projects/aracena_modeling/Inputs/counts_matrices/final_testing/fully_preprocessed_flu_expression_for_predictDBNORMSTRAT.txt",
"NI":"/beagle3/haky/users/saideep/projects/aracena_modeling/Inputs/counts_matrices/final_testing/fully_preprocessed_NI_expression_for_predictDBNORMSTRAT.txt"}
window_sizes = [4]
for normstrat in normalization_strategies:
for context in contexts.keys():
for window_size in window_sizes:
cur_proj_dir = os.path.join(path_to_project_dir,context+"_ws"+str(window_size)+"_ns"+normstrat)
os.makedirs(cur_proj_dir,exist_ok=True)
with open(path_to_config_template,"r") as f:
config = json.load(f)
config["general_parameters"]["project_directory"] = cur_proj_dir
config["general_parameters"]["context"] = context
config["generate_enpact_training_data"]["input_files"]["normalized_expression_data"] = contexts[context].replace("NORMSTRAT",normstrat)
config["generate_enpact_training_data"]["reference_epigenome"]["num_bins"] = window_size
with open(os.path.join(cur_proj_dir,"config.json"),"w") as f:
json.dump(config,f, indent=4)
with open(path_to_run_linearization_template,"r") as f:
run = f.read()
run = run.replace("CONFIG_FILE",os.path.join(cur_proj_dir,"config.json"))
run = run.replace("JOBNAME",context+"_ws"+str(window_size))
run = run.replace("ERROR_LOG", os.path.join(cur_proj_dir,"error_linearization.log"))
run = run.replace("OUTPUT_LOG", os.path.join(cur_proj_dir,"output_linearization.log"))
with open(os.path.join(cur_proj_dir,"run_linearization.sbatch"),"w") as f:
f.write(run)
subprocess.run(["sbatch",
os.path.join(cur_proj_dir,"run_linearization.sbatch")],
cwd=cur_proj_dir)Submitted batch job 18644266
Submitted batch job 18644267runtime: ~ 1 hour per TWAS run (SPrediXcan)
path_to_run_twas_template = "/beagle3/haky/users/saideep/github_repos/Con-EnPACT/run_twas.sbatch"
path_to_project_dir = "/beagle3/haky/users/saideep/projects/Con_EnPACT/models"
for normstrat in normalization_strategies:
for context in contexts.keys():
for window_size in window_sizes:
cur_proj_dir = os.path.join(path_to_project_dir,context+"_ws"+str(window_size)+"_ns"+normstrat)
os.makedirs(cur_proj_dir,exist_ok=True)
with open(path_to_run_twas_template, "r") as f:
run = f.read()
run = run.replace("CONFIG_FILE",os.path.join(cur_proj_dir,"config.json"))
run = run.replace("JOBNAME",context+"_ws"+str(window_size))
run = run.replace("ERROR_LOG", os.path.join(cur_proj_dir,"error_twas.log"))
run = run.replace("OUTPUT_LOG", os.path.join(cur_proj_dir,"output_twas.log"))
with open(os.path.join(cur_proj_dir,"run_twas.sbatch"), "w") as f:
f.write(run)
subprocess.run(["sbatch",
os.path.join(cur_proj_dir,"run_twas.sbatch")],
cwd=cur_proj_dir)Submitted batch job 18680959
Submitted batch job 18680960