Code
import os,sys
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as snsSaideep Gona
March 24, 2024
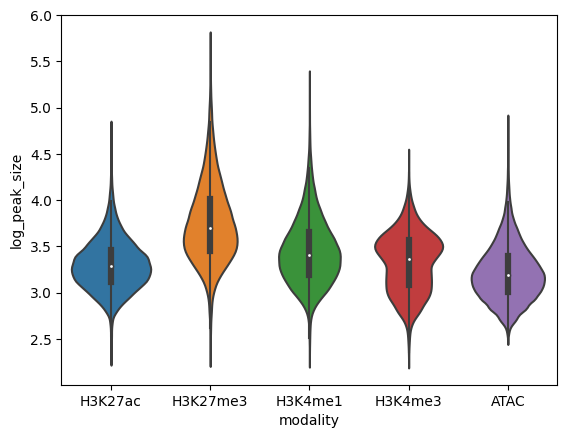
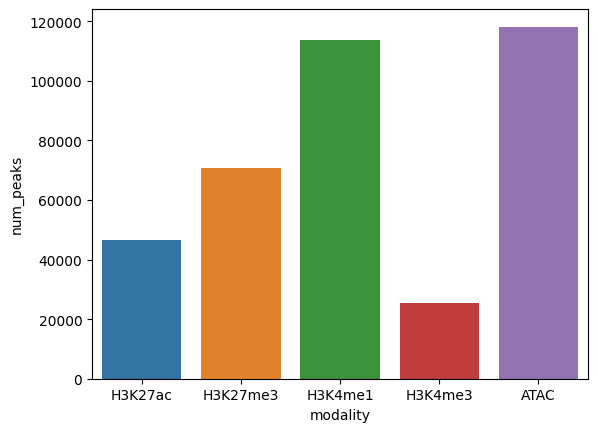
Peak data has a number of differences from expression data. Things like the size of peaks relative to genes, number of peaks in a sample, peak overlap between modalities, are helpful to assess in order to make EnPACT models.
peak_sizes = []
modality_match_vec = []
modalities_in_order = []
peak_counts = []
for modality in modality_pcs.keys():
cur_count_file = os.path.join(path_to_count_table,f"fully_preprocessed_{modality}_{modality_pcs[modality]['Flu']}_{modality_pcs[modality]['NI']}.txt")
all_expression_ori = pd.read_csv(
cur_count_file,
sep=" ")
peak_counts.append(all_expression_ori.shape[0])
modalities_in_order.append(modality)
cur_peak_sizes = []
for region in all_expression_ori.index:
if len(region.split("_")) != 3:
continue
peak_sizes.append(int(region.split("_")[2])-int(region.split("_")[1]))
modality_match_vec.append(modality)
peak_sizes_for_plot = pd.DataFrame({"peak_size":peak_sizes, "log_peak_size":np.log10(peak_sizes) ,"modality":modality_match_vec})
sns.violinplot(x="modality",y="peak_size",data=peak_sizes_for_plot)
plt.show()
sns.violinplot(x="modality",y="log_peak_size",data=peak_sizes_for_plot)
plt.show()
num_peaks_for_plot = pd.DataFrame({"modality":modalities_in_order, "num_peaks":peak_counts})
sns.barplot(x="modality",y="num_peaks",data=num_peaks_for_plot)
plt.show()
print("Minimum peak size", min(peak_sizes))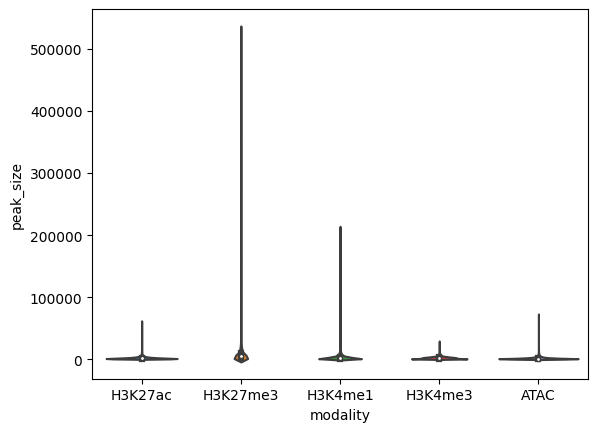
Minimum peak size 183